Myoglobin
| 
|
|
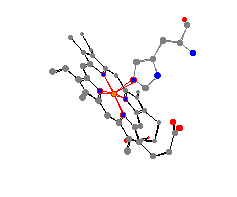
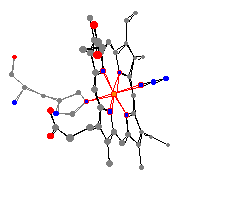
As can be seen from the views produced by the program,
the geometry of deoxymyoglobin is square pyramidal, while the
geometry of myoglobin with azide is octahedral. This geometry,
along with the bond angles and distances between the iron and the
surrounding atoms can be seen especially well with the paper models
produced by the program. These models allow us to make comparisons
between the distances, and thus the strength of bond, between the iron
and the surrounding molecules when the ligand is and is not present.
| For example, we find out from these models that overall
the nitrogens in the heme group are held much more closely in the
deoxymyoglobin. (The average deoxymyglobin distance is 190 pm versus
202 pm for myoglobin with the azide.) This can be explained, as the N3
in 1swm should lead to a higher electron
density around the iron, and an overall more shielded iron center, thus
increasing the size of the heme pocket.
| |
Hemoglobin
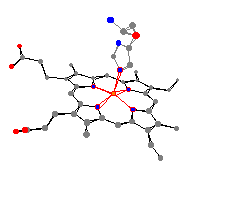
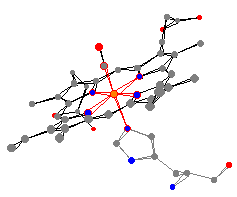
ligands using the paper models. The same general conclusion can
be drawn
as the one drawn for myoglobin. The heme pocket gets larger when
a ligand is added.
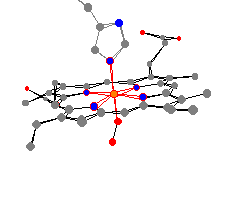
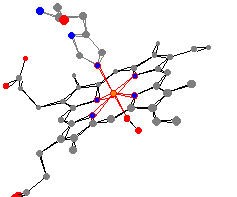
Here are the two most important myoglobin and
hemoglobin molecules. These are the oxy-proteins, where an
O2 is bound to the iron across from the histidine.
It is possible to use Molecular Origami again to make comparisons
between the bond angles and distances of the two proteins.
For example, the average nitrogen heme bond distances
are very similar (199 pm for hemoglobin and 200 pm for myoglobin).
Iron-Sulfur Systems
Two Other Interesting Proteins