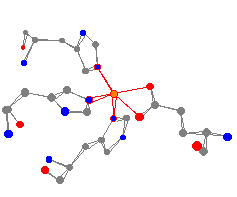
| As can be seen from this view of the protein, the iron is bonded to three histidine groups (in the usual manner through a nitrogen in the ring) and one glutamic acid carboxylate. | |
This iron center has a fairly distorted square pyramidal geometry.
The average N-Fe bond is 219 pm and the two oxygen atoms are at 185
and 214 pm.2
| |
2 Using the default bond distance of 250 pm, there was a bond drawn
between the carbon and iron atoms, which are 226 pm apart. Since there is
not actually a bond to the carbon in the glutamic acid, the bond was removed
by setting the maximum bond distance to 220 pm. When this is done we get the
more correct bonding picture of the molecule where the only atoms from the glutamic
acid in the foldable Molecular Origami model are the two oxygens.
This interesting view is the iron in the photosynthetic reaction center
of a bacterium. The reaction center controls the primary processes of
photosynthesis, the changing of energy from electromagnetic radiation
into chemical energy.
Photosynthetic Reaction Center
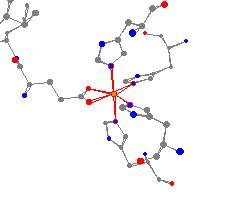
As can be seen in the view, the iron is surrounded by four histidine
molecules
and one glutamic acid. The geometry of this iron is different from
the iron in transferase because of
the larger number of histidines, and could best be described as a
distorted
octahedron.3 Again, valuable information could be obtained by comparing
the angles
and distances of the bonds from the iron to the two atoms in the
glutamic acid.
The axial nitrogens are at an
average of 232 pm,
while the equatorial nitrogens have an average distance of 214 pm. The
oxygens are 212
and 215 pm from the iron atom. The iron-ligand bond distances are slightly longer than
the above transferase.
This can be explained, as the extra histidine in 1aig should lead to a higher electron
density around the iron, and an overall more shielded iron center.
this default distance is lowered to below 244 pm, the program will draw a
bond between those two atoms. For this view, the value was set to 242 pm.
Isolated Non-Heme Iron
Iron-Sulfur Systems