Molecular Origami |
 |
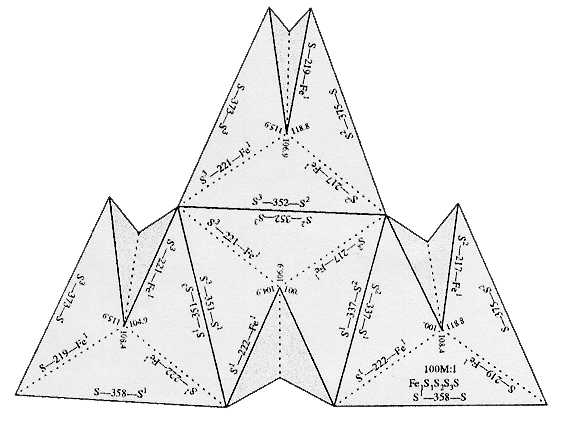 |
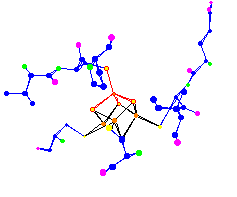
| [4Fe-4S] Ferredoxin
Active Site |
Click to enlarge and sharpen |
CoolMolecules: A Molecular Structure Explorer
Iron and Zinc in Enzymes
Introduction to Molecular Geometry
National American Chemical Society Presentation, San Diego, California, April 4, 2001
A Small-Molecule and Metalloprotein Searchable Structural Database
Model Gallery
Hydroxyapatite Model
Molecular Origami - the concept
Molecular Origami is an outgrowth of 'Data-Driven Chemistry', a technique
of teaching chemistry under development by Prof. Bob Hanson at
St. Olaf College, in Northfield, Minnesota.
Molecular Origami refers to the detailed folding and arrangement of
atoms in molecules and network solids. Molecular
Origami by Robert M. Hanson (University Science Books) is a set of
actual paper models focusing on small organic and inorganic structures.
Data are based primarily on electron and x-ray diffraction results.
Discussion of the models in terms of bonding and qualitative molecular
orbital theory is included. Exact interatomic distances and angles are
indicated on these figures. They can be folded to produce
three-dimensional scale models of molecules or sections of molecules.
Molecular Origami - the PC program DOWNLOAD
The program Molecular Origami allows you to print most of the
structures in the book. In addition, you can
investigate molecular structure on your own, using data that you might
have access to. The program has an option to retrieve data directly
from the internet. Some databases that can be freely accessed include:
Brookhaven's Protein Data Bank, SWISS-MODEL Repository, and Indiana
University Molecular Structure Center.
There is not a Mac Version of Molecular
Origami.
Get a taste of the program by looking at:
Why Use Molecular Origami ?
The computer program originated with the idea of producing paper scale
models of proteins. As we looked for other protein computer modelers it
became evident that the real value of the program was not just the
origami, but also the computer modeling capabilities. Other protein
modeling programs exist such as RasMol. For a task such as showing the
entire protein structure RasMol does a great job. In fact if you have
RasMol, Molecular Origami has a button that displays the current protein
with RasMol. Molecular Origami is unique because it enables you to pick
and choose what groups you want to look at. This is useful because many
proteins have hundreds
of amino acids and get extremely complicated to view in their entirety.
Click here to see an entire protein modeled by
RasMol.
Click here to see the same protein modeled by
Molecular Origami
With Molecular Origami you can view only the parts of the protein you are
interested in. It is easy, all you have to do is select the groups you
want to appear from a list. They appear on the screen where they can be
rotated by moving the mouse.
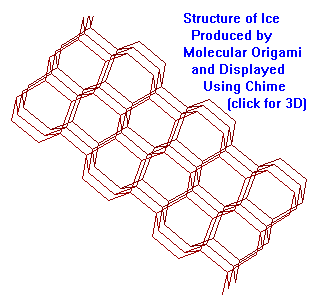
Unique capabilities of Molecular Origami include:
- Direct search and download of data from the internet
- Ability to select which groups you want to view
- Create 3-D active
Chime files for export to Netscape
- Direct link to
Rasmol for full-protein viewing
- Ability to save up to 9 views with each view able to show different
groups within the protein
- Ability to work with Unit Cells using crystallographic information.
- Ability to look at common geometries using internal coordinates.
- Easy mouse-following rotation
Using Molecular Origami
Information Related to Molecular Origami
Origami & Protein Related Links
X-ray Crystallography Information
Origami.EXE is free to download for PC as origam##.zip where ## is the
version.
Modular Origami
Questions? Comments? Suggestions?
E-MAIL
Prof. Hanson
Ben Murray

Disclaimer


